Introduction
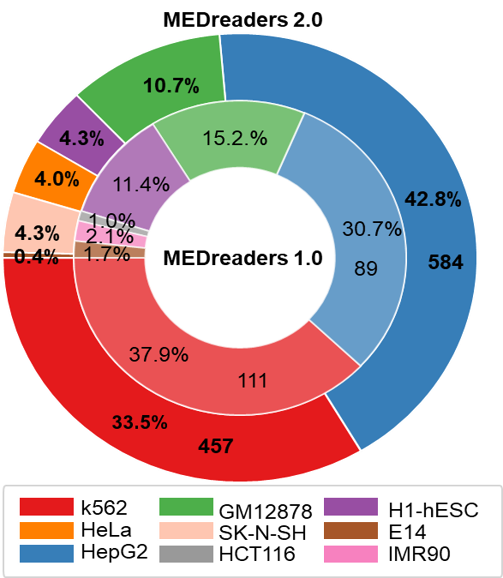
MeDReaders 2.0 is an upgrade to the original MeDReaders database and webserver. The new MeDReaders 2.0 covers interactions between transcription factors (TFs) and DNA modifications involved in methylation and active demethylation, such as hydroxymethylation (hmC), formylation (fC), and carboxylation (caC). This update adds 257 mC-DNA, 160 hmC-DNA, 220 fC-DNA, and 107 caC-DNA binding activities.
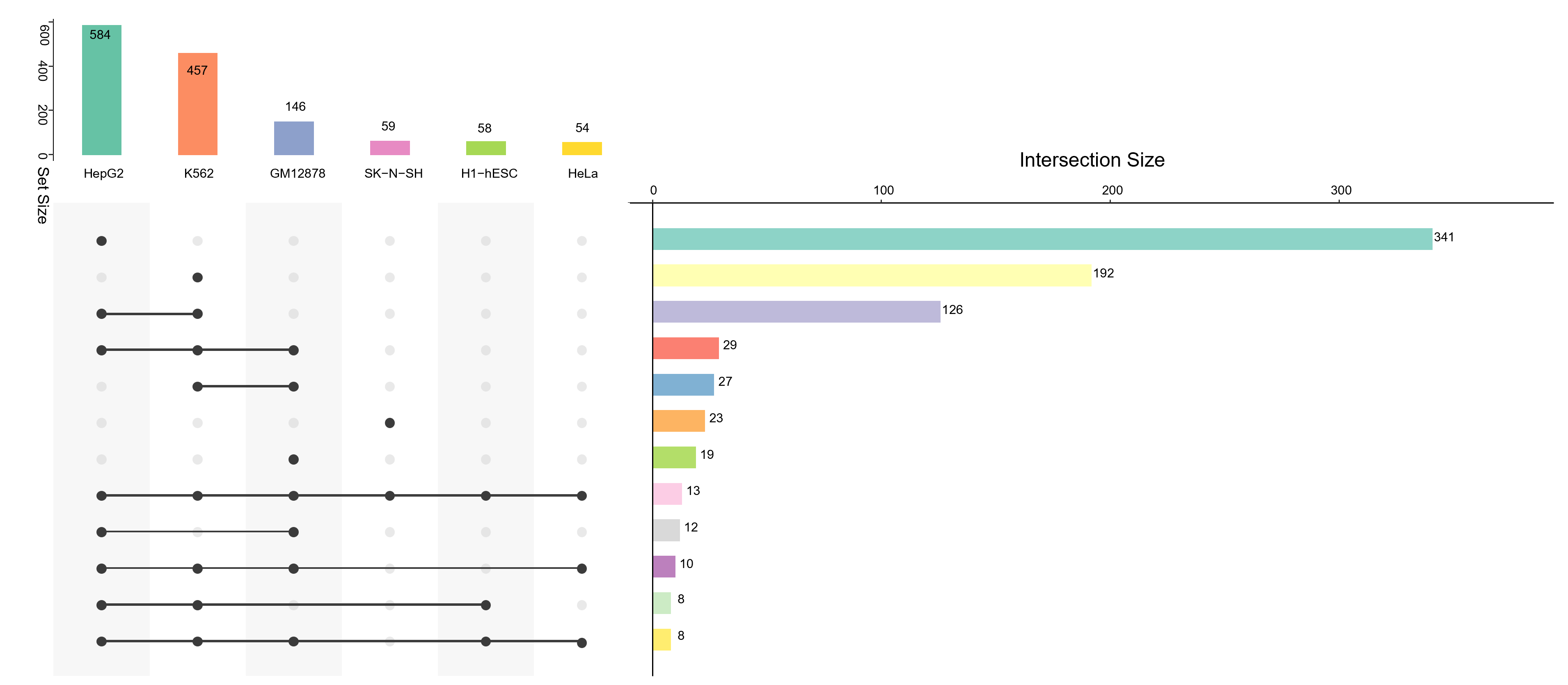
A total of 774 TFs that bind to methylated DNA sequences were manually curated from human and mouse studies reported in the literature. In silico approaches predicted methylated and unmethylated motifs for 1,363 TFs by integrating whole genome bisulfite sequencing (WGBS) and ChIP-Seq datasets from six human cell lines and one mouse cell line, extracted from ENCODE and NCBI databases.
MeDReaders 2.0 thus provides a more comprehensive resource by covering epigenetic modifications of three methylated oxides, offering valuable data for further studies and aiding in experimental design related to the dynamic regulation of DNA methylation and active demethylation.
TFs Obtained through Literature RetrievalTranscription factors summarized from published literatures for "High-methly(TFs)" page:
| Species | No. of TFs | No. of Cells/Tissues |
|---|---|---|
| Human | 601 | 4 |
| Mouse | 130 | 4 |
Transcription factors summarized from published literatures for "Multiple-modification (TFs)" page:
| Experiment | No. of TFs | No. of Modification Type |
|---|---|---|
| DAPPL | 332 | 4 |
| MS | 437 | 3 |
- achieve methylated DNA binding motifs of TFs of user's interest or search potential TFs by matching methylated binding sequences as user requested.
- obtain and/or compare methylation contexts of the same TF under different cell lines and/or tissues.
- download position weight matrix (PWM) of TFs associated with methylation status (high or low methylation).
- submit newly published information of methylated-DNA binding TFs to enrich our database.
Release and Version Information
Jun. 1, 2024, MeDReaders 2.0: A Database for Modified DNA Readers v2.0 was released.
Jul. 1, 2017, MeDReaders: A database for Methylated DNA Readers v1.0 was released.
Contact
Guohua Wang, professor
Northeast Forestry University, Harbin 150040, China
Mail: ghwang@nefu.edu.cn
Statistics
| Readers v1.0 vs Readers v2.0 | Multiple modification distribution |
|---|---|
 |
 |
| Human | Mouse |
|---|---|
 |
E14: 5 |