PlantCADB
Plant Chromatin Accessibility Database
What is PlantCADB?
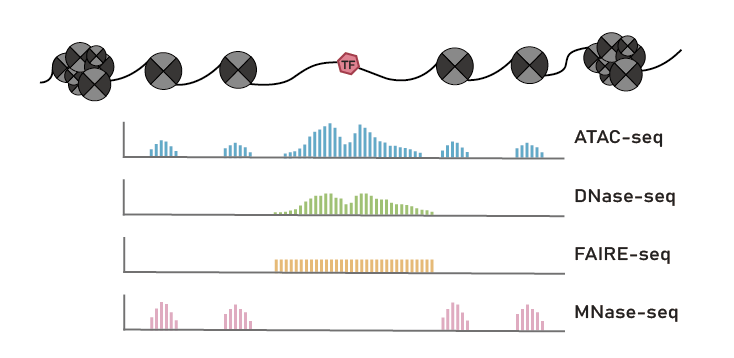
Chromatin accessibility landscapes are essential to detecting regulatory elements, illustrating the corresponding regulatory networks and, ultimately, understanding genetic bases underlying key biological processes.
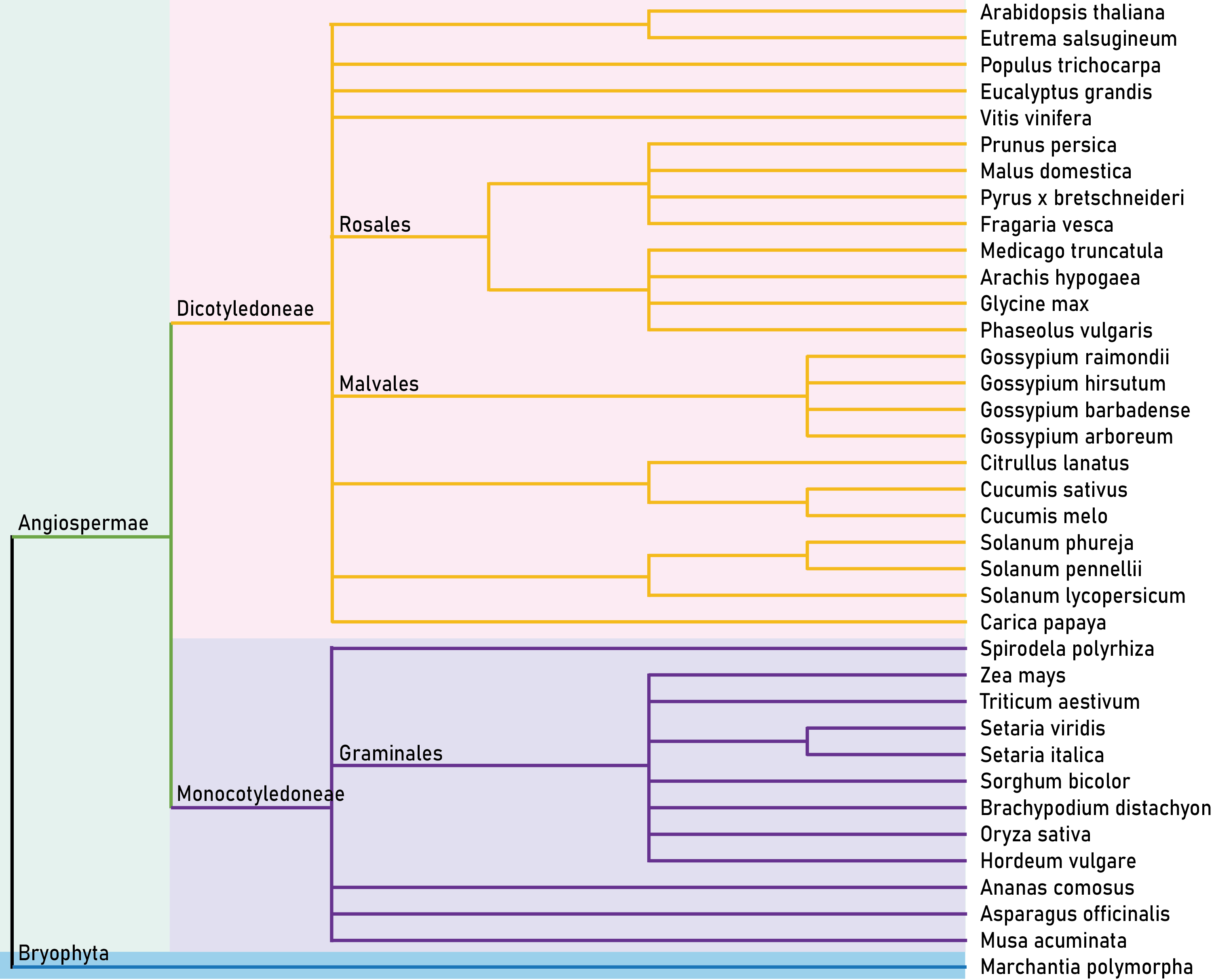
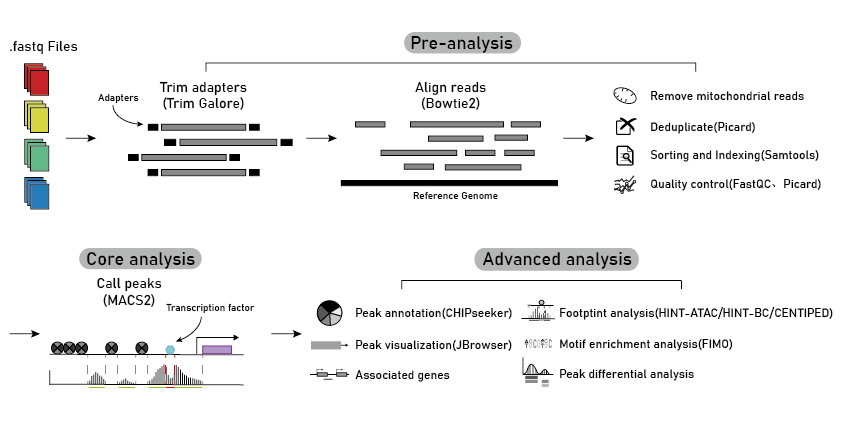
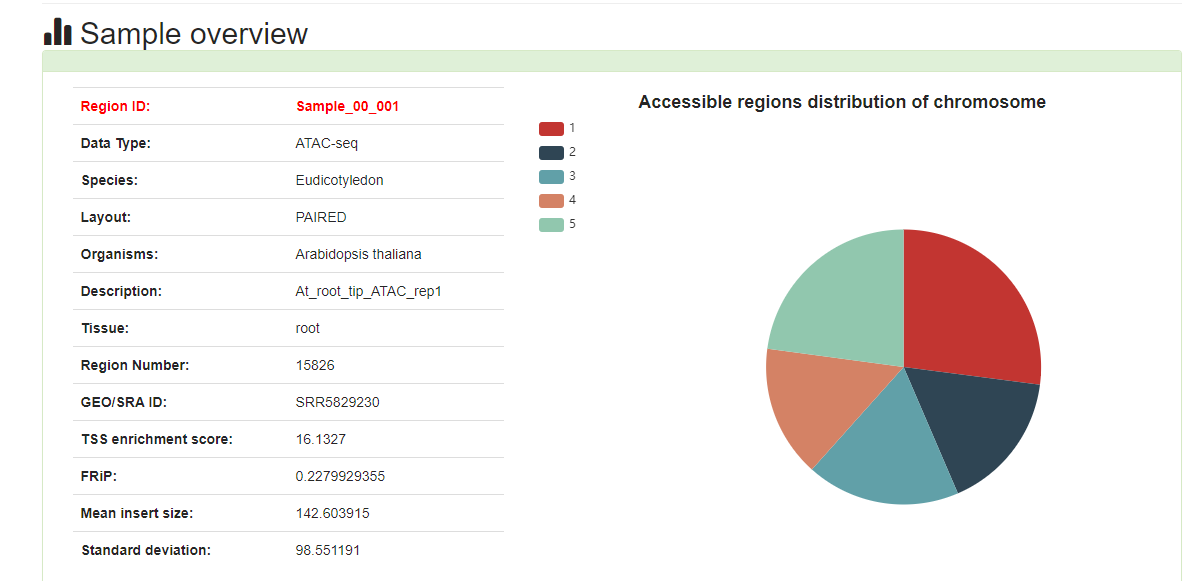
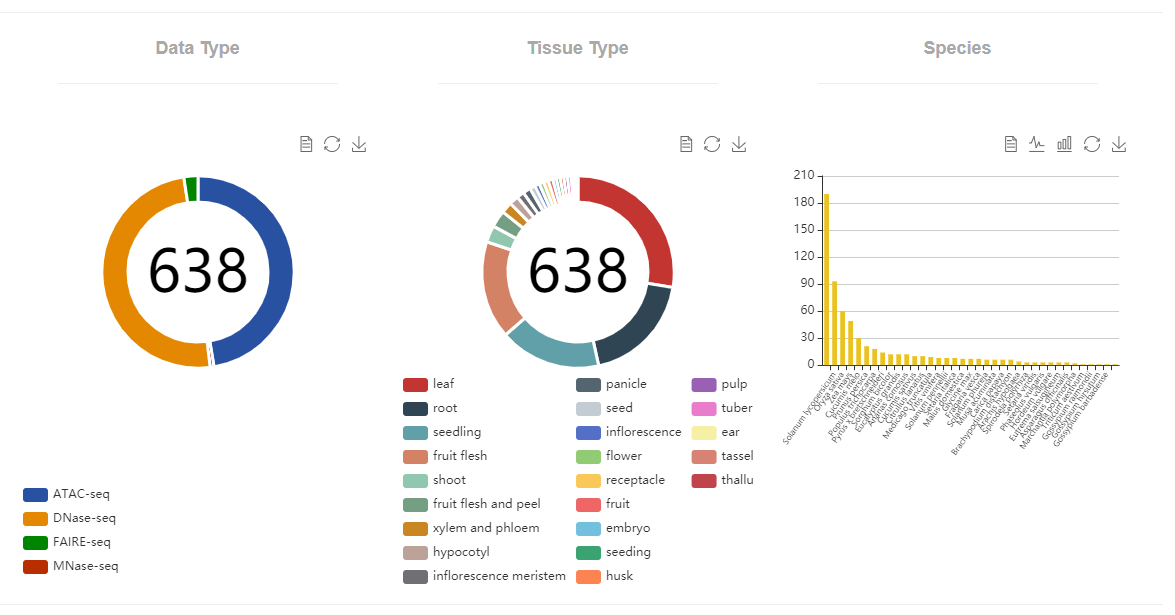
Here, we developed a comprehensive plant chromatin accessibility database (PlantCADB, https://bioinfor.nefu.edu.cn/PlantCADB/) to advance the understanding of molecular networks regulating plant development, physiology and stress adaptation. It contains 18,339,426 accessible chromatin regions (ACRs) from more than 600 samples of 37 species. These ACRs were annotated with genomic information, associated genes, transcription factors footprint, motif and SNPs. Additionally, PlantCADB provides various tools to visualize ACRs and corresponding annotations. It thus forms an integrated, annotated and analyzed plant-related chromatin accessibility information which can aid to better understand genetic regulation networks underlying development, important traits, stress adaptions and evolutions.
| Type | Species | ATAC-seq | DNase-seq | FAIRE-seq | MNase-seq | ||
|---|---|---|---|---|---|---|---|
| GEO | SRA | GEO | SRA | SRA | SRA | ||
| Dicotyledoneae 471 |
Arabidopsis thaliana | 85 | 27 | 48 | 12 | 15 | 3 |
| Solanum lycopersicum | 16 | 77 | |||||
| Cucumis melo | 30 | ||||||
| Prunus persica | 21 | ||||||
| Populus trichocarpa | 18 | ||||||
| Pyrus x bretschneideri | 14 | ||||||
| Eucalyptus grandis | 12 | ||||||
| Citrullus lanatus | 10 | ||||||
| Cucumis sativus | 10 | ||||||
| Medicago truncatula | 9 | ||||||
| Solanum pennellii | 8 | ||||||
| Vitis vinifera | 8 | ||||||
| Glycine max | 7 | ||||||
| Malus domestica | 7 | ||||||
| Fragaria vesca | 7 | ||||||
| Solanum phureja | 6 | ||||||
| Carica papaya | 6 | ||||||
| Arachis hypogaea | 4 | ||||||
| Eutrema salsugineum | 3 | ||||||
| Phaseolus vulgaris | 3 | ||||||
| Gossypium arboreum | 1 | ||||||
| Gossypium barbadense | 1 | ||||||
| Gossypium hirsutum | 1 | ||||||
| Gossypium raimondii | 1 | ||||||
| Monocotyledon 165 |
Zea mays | 23 | 13 | 11 | 2 | ||
| Oryza sativa | 58 | 2 | |||||
| Sorghum bicolor | 3 | 3 | 6 | ||||
| Ananas comosus | 12 | ||||||
| Setaria italica | 2 | 6 | |||||
| Brachypodium distachyon | 3 | 3 | |||||
| Musa acuminata | 6 | ||||||
| Hordeum vulgare | 3 | ||||||
| Setaria viridis | 3 | ||||||
| Asparagus officinalis | 3 | ||||||
| Spirodela polyrhiza | 3 | ||||||
| Triticum aestivum | 1 | ||||||
| Hepaticae 2 | Marchantia polymorpha | 2 | |||||