Recent Publications
近期文章
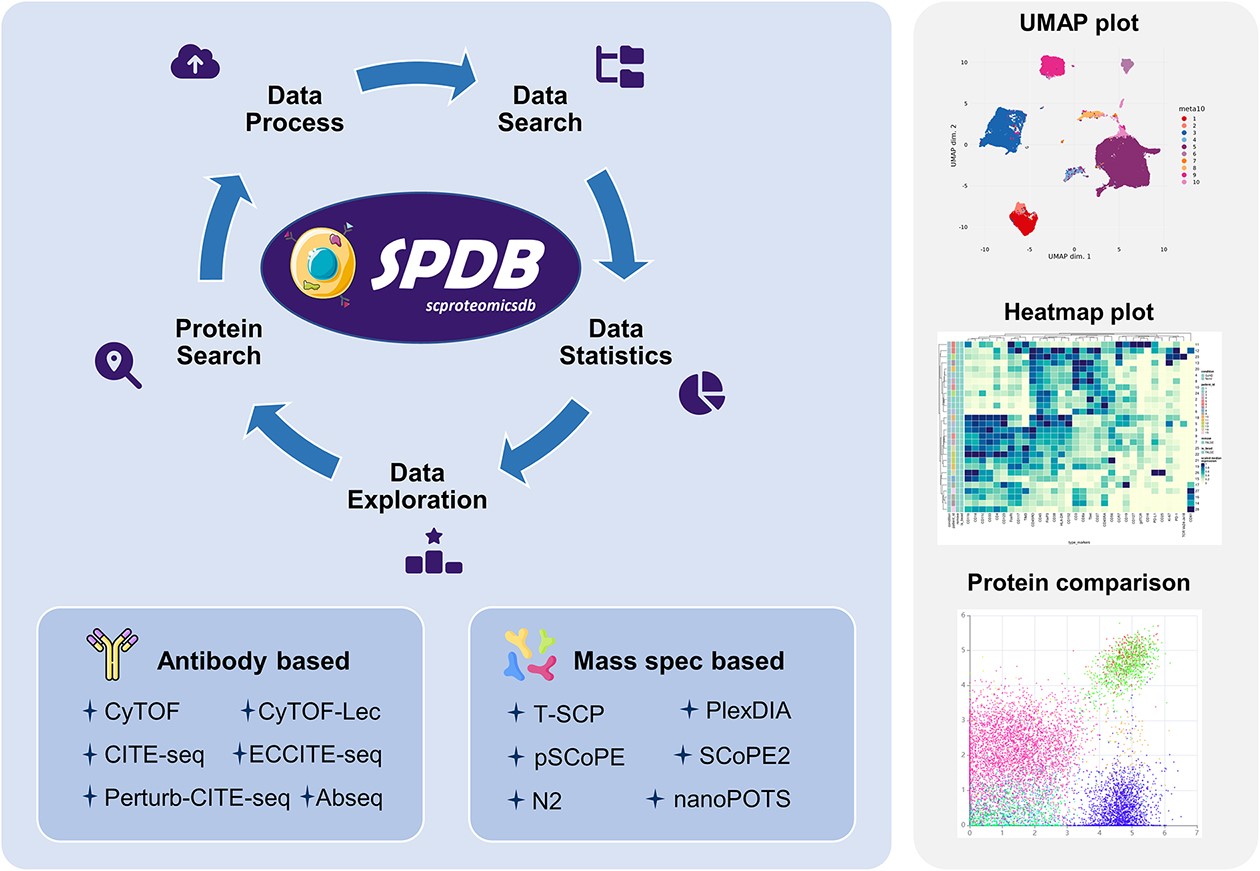
SPDB: a comprehensive resource and knowledgebase for proteomic data at the single-cell resolution.
Fang Wang, Chunpu Liu, Jiawei Li, Fan Yang, Jiangning Song , Tianyi Zang, Jianhua Yao*, Guohua Wang*.
Nucleic Acids Research, 2024.
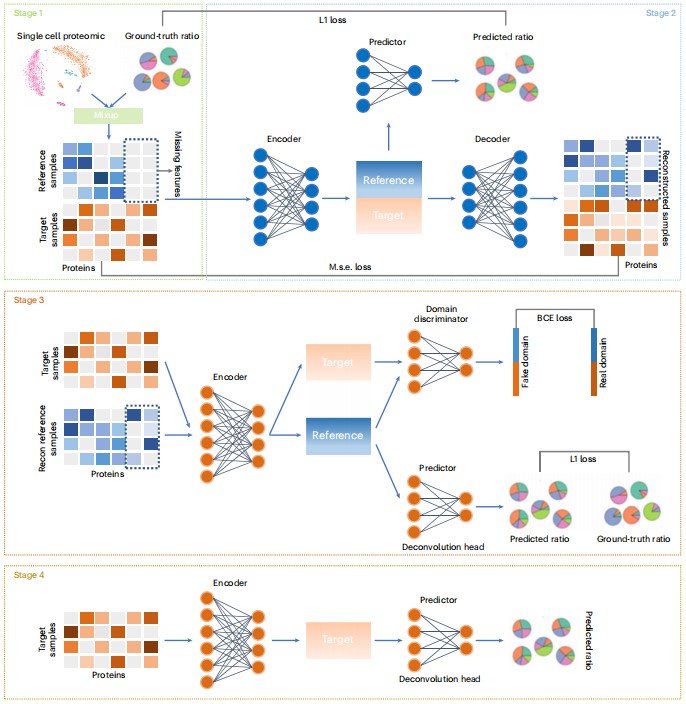
Deep domain adversarial neural network for the deconvolution of cell type mixtures in tissue proteome profiling.
Fang Wang, Fan Yang, Longkai Huang, Wei Li, Jiangning Song, Robin B. Gasser, Ruedi Aebersold, Guohua Wang & Jianhua Yao.
Nature Machine Intelligence, 2023.
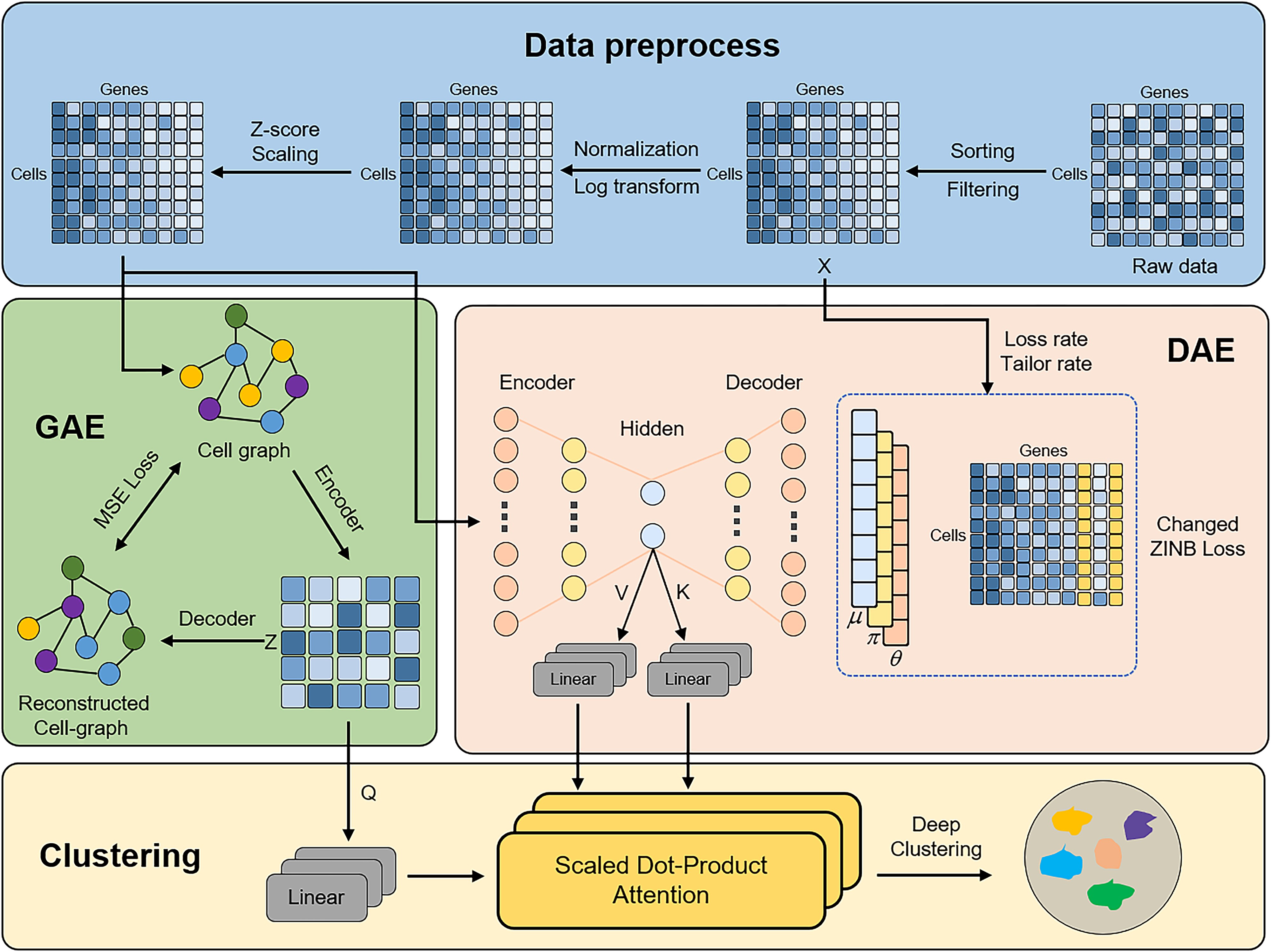
scLEGA: an attention-based deep clustering method with a tendency for low expression of genes on single-cell RNA-seq data.
Zhenze Liu, Yingjian Liang, Guohua Wang, Tianjiao Zhang.
Briefings in Bioinformatics, 2024.
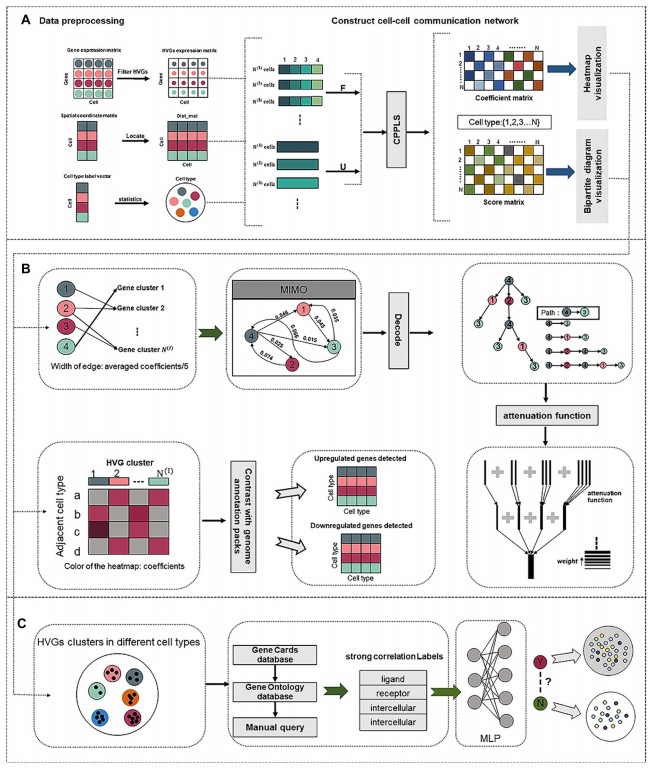
CPPLS-MLP: a method for constructing cell-cell communication networks and identifying related highly variable genes based on single-cell sequencing and spatial transcriptomics data.
Tianjiao Zhang, Zhenao Wu, Liangyu Li, Jixiang Ren, Ziheng Zhang, Guohua Wang*.
Briefings in Bioinformatics, 2023.